A Few Thoughts on Troubleshooting of DNA Sequencing - Part II
In the first part we discussed how to troubleshoot no or low signals. Basically empty electropherograms. Very frustrating. Now, I focus on another frequent result – you obtain some signals but the height of the DNA sequencing peaks diminished rapidly. A read length is very short.
A Very Short Read Length
The height of the DNA sequencing peaks always diminish (die out) slightly with distance from the primer, this is quite normal and best seen in the Raw data. I do not recommend the electropherogram to monitor this decline because there are various height normalization procedures available in the data analysis software used to process your samples and consequently depending on the basecalling algorithm we use to analyze your data you can observe the decline in the electropherogram but you may not. Please note that if you are troubleshooting your results, you can neglect electropherograms in some cases but you should never neglect RAW data!
Anyway, if the peak height dies out more quickly then one would wish it makes the sequence readable only at its beginning, giving rise to poor quality basecalling towards the end of the sequence because the signal:noise ratio decreases and the basecalling algorithm is simply unable to read the sequence properly.
Unfortunately as in other cases, also this symptom has different causes.
Improper ratio of template to primer in the sequencing reaction
Primers in sequencing reactions are used at 5 micromolar. Using more primer can cause mis-priming and mixed sequence. If, on the other hand, the primer concentration is optimal but DNA amount is too high, more than optimal priming events will occur. This consequently means that dNTPs are used up quicker which increases the termination rate. This implies that the amount of DNA and primer in a sequencing reaction will affect how long of a readable sequence one can obtain.
Too much DNA in the sequencing reaction
This is fairly the same problem as improper ratio of template to primer described above. Too much DNA causes the sequence to die out very quickly or even fail totally. dNTPs are used up fast and only short products are observed. Additionally, too much DNA means too much phosphate (in its backbone) chelating magnesium ions from the reaction mixture.
Homopolymers or repeats
Die out sometimes also occurs when trying to sequence through homopolymer regions or repeats but this will be discussed in the last part of this miniseries.
Salts and other inhibitors in the sequencing reaction
We discussed this problem already in the first chapter. The presence of inhibitors, most frequently salts but also ethanol, phenol, etc. has a strong influence on polymerase processivity during sequencing reaction and it can kill the reaction completely. If their concentration is low you will get some signal but diminishing very early.
We have run two reactions to show you this effect – the picture below shows two samples differing only in NaCl concentration, there is no other difference there. Please note the decline of signal in one of electropherograms! It speaks for itself.
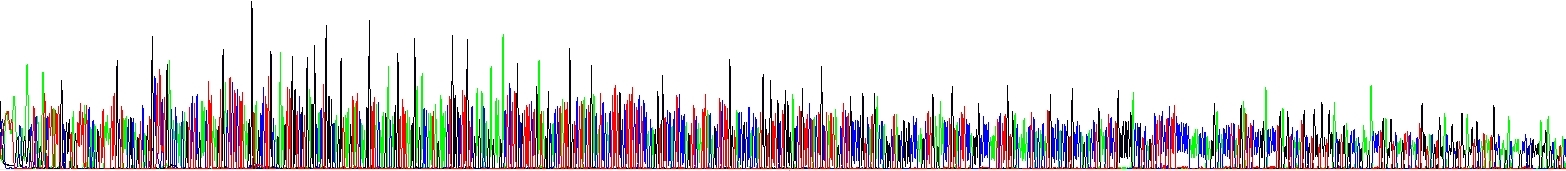
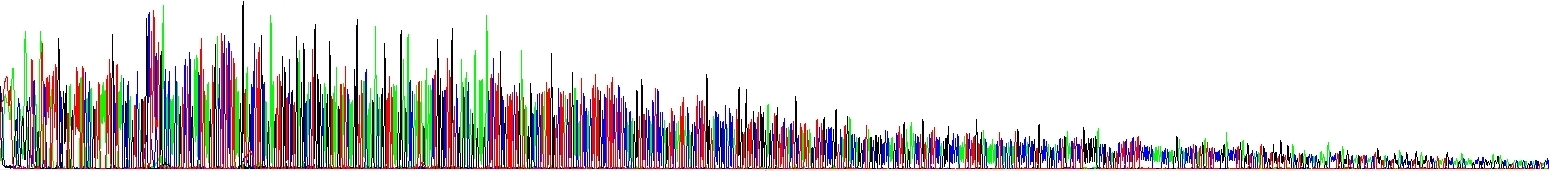
Signal Die Out due to hairpins
Certain unusual DNA sequences will inhibit the Taq polymerase, hairpins being the most prominent of them. To sequence through these regions is a quite challenging scenario. First, they are naturally stable. Second, cycle sequencing kits contain dITP instead of dGTP to reduce secondary structures during electrophoresis and thus allowing better resolution (guanine forms three hydrogen bonds whereas inosine only two). I mean, for most samples it is good to use dITP. But the disadvantage of using dITP is that it is not incorporated as efficiently as dGTP by the DNA polymerase. Hairpins impede the DNA polymerase themselves and even more if using dITP. The resulting slow down of the polymerase causes the peak height of the sequencing products to decrease, the more stable the DNA fold, the bigger the decrease in peak height.
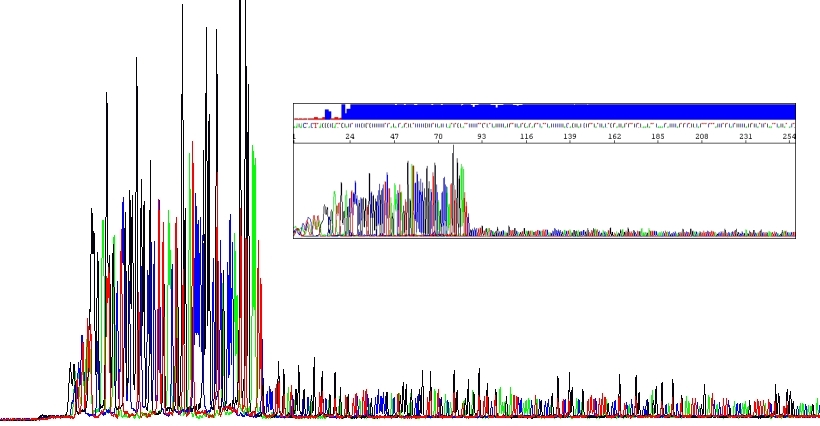
One can think of bringing dGTP back to the game but this is usually of no or little help and we do not offer this option. Instead, we can circumvent this problem by using analogues of dNTPs in the PCR reaction. If you are sequencing hairpins, use our HairpinSeq protocol which includes sequencing from both directions!
Sanger lab, info@seqme.eu