PCR products purification - kit comparison and useful tips
The aim of this test was to determine if and how the PCR purification kits have an effect on the quality of the Sanger sequencing outputs in our laboratory. The conclusions and recommendations below are not generally valid and it is recommended that everyone conduct a similar test using their own samples.
Why we tested
When sequencing PCR products, sometimes the entire sequence is not read:
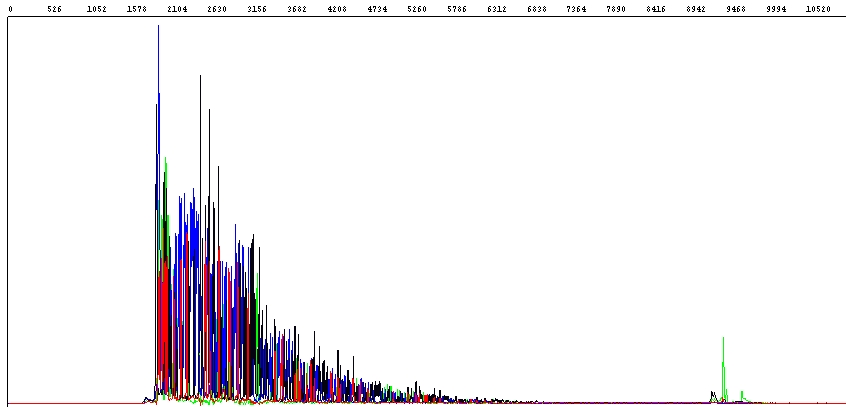
As you can see in the raw data in Figure 1 above, the fluorescence signal is initially very good, but drops sharply, and about half of the PCR product sequence is unreadable (the end of the PCR product is seen as a distinct green peak on the right).
A typical cause of this problem is the presence of various inhibitors in the sequencing reaction templates. These may be inhibitors coming from the original biological materials (tissue, blood, soil, etc.) that have not been removed during DNA isolation or inhibitors coming from DNA isolation or cleanup procedures (residues of binding and / or washing buffers - various salts, detergents, alcohols).
Methods for genomic DNA isolation are quite variable and depend on the type of input material. For Sanger sequencing, PCR amplification of the genomic DNA obtained and purification of the PCR product is often a typical next step, and it is relatively standardized. PCR products can be purified enzymatically, cut from gels, or by using cleanup columns, etc. In our test we focused on this last method - purification of PCR products on spin columns. Our aim was to find out how kits for purification of PCR products from different manufacturers influence the quality (purity) of the obtained DNA template and subsequent sequencing.
How we tested
Nowadays, you can choose from a plethora of PCR product purification kits, which differ in many details. The main differences are:
- column design (narrow / wide neck of silica membrane)
- centrifugation time
- RCF
- number of washing steps
- additional centrifugation step before elution
- elution time
- volume of elution buffer
For this test, we have selected four commonly used PCR product purification kits:
- NucleoSpin® Gel and PCR Clean-up, 740609.10, Macherey-Nagel, protocol version 02/2017 / Rev. 04, hereinafter referred to as Macherey-Nagel
- Monarch® PCR & DNA Cleanup Kit (5 µg), T1030G, New England Biolabs, protocol version 1.1-10.14, hereinafter referred to as NEB
- QIAquick PCR Purification Kit, 28104, QIAGEN, protocol version 07/2018, hereinafter referred to as Qiagen
- DNA Clean & Concentrator-5 Kit (TM), D4003S, ZYMO RESEARCH, protocol version 1.2.1., hereinafter referred to as Zymo
Comparison of the main protocol parameters of selected kits (Table A):
| Kit | RCF | Centrifugation time | Number of washing steps | Washing buffer volume (ul) |
Additional centrifugation before elution |
Incubation time with elution buffer | Elution buffer volume (ul) |
|
Macherey-Nagel
|
11000
|
30 sec (1 min after elution)
|
2x
|
700
|
1 min
|
1 min
|
15-30
|
|
NEB
|
16000
|
1 min
|
2x
|
200
|
optional
|
1 min
|
>=6
|
|
Qiagen
|
17900
|
30-60 sec (1 min after elution)
|
1x
|
750
|
1 min
|
1 min
|
30-50
|
|
Zymo
|
>=10000
|
30 sec
|
2x
|
200
|
no
|
1 min
|
>=6
|
For the purposes of our test, we used the following protocols (Table B):
| Kit | RCF | Centrifugation time | Number of washing steps | Washing buffer volume (ul) | Additional centrifugation before elution | Incubation time with elution buffer | Elution buffer volume (ul) |
|
Macherey-Nagel
|
According to the protocol |
20
|
|||||
|
NEB
|
According to the protocol |
no
|
According to the protocol |
15
|
|||
|
Qiagen
|
16000*
|
1 min
|
According to the protocol |
30
|
|||
|
Zymo
|
16000
|
According to the protocol |
15
|
||||
* Max. RCF value of our centrifuge is 16000.
The same PCR products (different lengths from 150 to 750 bp) were always purified by different kits according to the protocols shown in Table B. The same amount of PCR product was applied to the columns. The quantitative yield (measured by Qubit 4 Fluorometer) was recalculated so that the same amount of DNA always entered the sequencing reaction. Therefore, the possible differences in the quality of the sequencing data could not be due to different amounts of the template in the sequencing reaction. However, this does not apply to the input template volume - for kits with lower yield and / or higher elution volume, naturally a higher template volume (and thus potentially more inhibitors) was used in the sequencing reaction. The purified PCR products were unidirectionally sequenced using the StandardSeq protocol. The quality of sequencing data was evaluated by simply looking at raw data and electropherograms. The evaluation was performed by three operators independently in Sequencing Analysis software (Applied Biosystems).
Results:
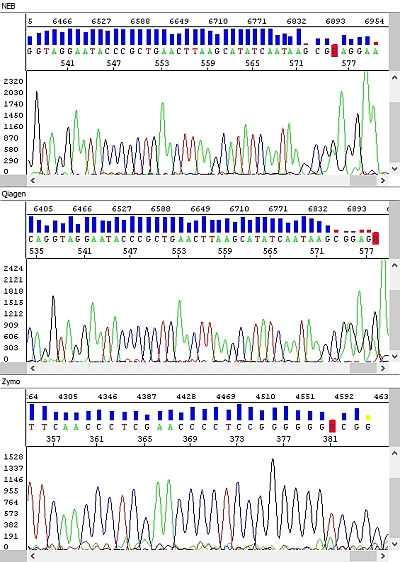
When comparing the data we found significant differences. We have selected a few examples below where these differences are clearly visible. All figures show the result of sequencing analysis of an identical PCR product (about 600 bp in length) purified by different kits.
Figure 2 - Raw data - top-down windows: NEB, Qiagen, Zymo, Macherey-Nagel:
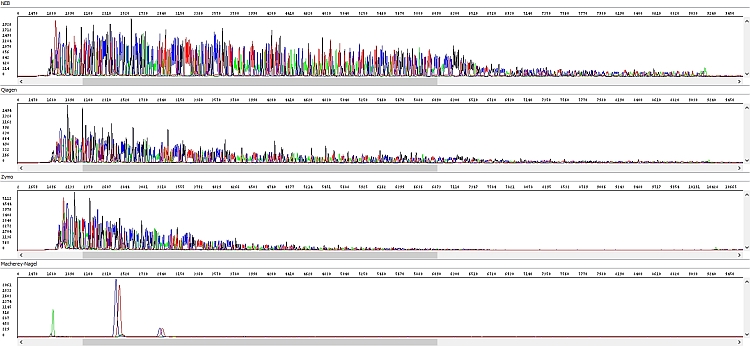
Figure 3 - Raw data, detail, view of the end of PCR product sequence - top-down windows NEB, Qiagen, Zymo:
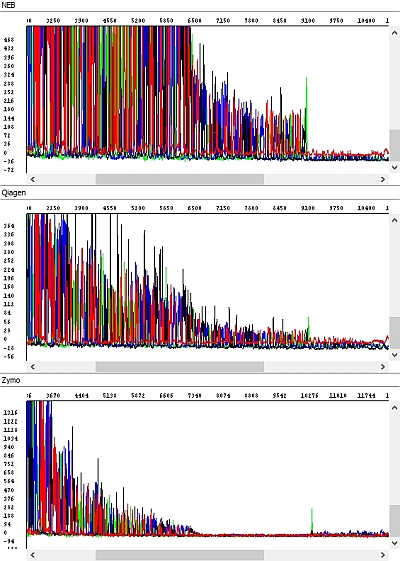
Figure 4 - Electropherograms - top-down windows: NEB (reading length QV20 559 bp), Qiagen (reading length QV20 559 bp), Zymo (reading length QV20 361 bp), Macherey-Nagel (reading length QV20 18):
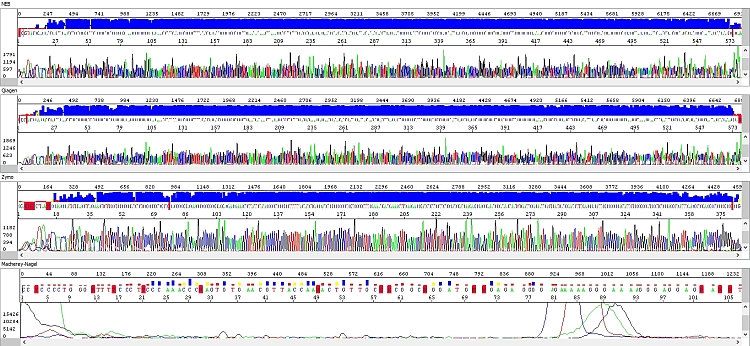
Figure 5 - Electropherograms, detail, view of the end of PCR product sequence - top-down windows: NEB, Qiagen, Zymo:
Comments on the results
The PCR products purified by the NEB kit were always without any problem, with high quality data along the entire length of the sequence (a possible decrease in the fluorescent signal was more related to the nature of the template - combination of individual bases, secondary structure, etc.)
Similar results were obtained with the Qiagen kit and although the signal decreased more rapidly in longer fragments (see raw data), it was still sufficient for sequencing analysis. However, we believe that if the PCR product concentration is below optimum, the Qiagen kit may not complete the sequencing of the PCR product, which would probably only happen with the NEB-purified PCR product with a really low concentration. The requirement for a higher volume of elution buffer and the above-mentioned consequences may also have an effect. In any case, in terms of our template concentration requirements, the Qiagen kit is perfectly ok.
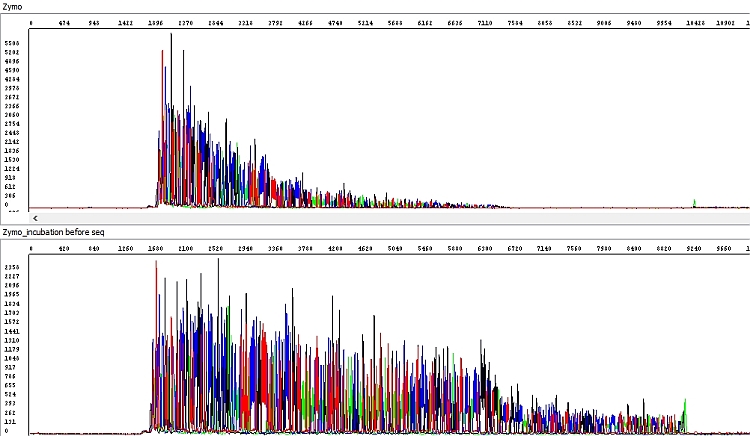
In the Zymo kit, we noted a significant deterioration in the quality of the sequencing data - the length of the sequencing reading was about half, and the raw data clearly show the absence of detectable peaks at the end of the sequence except for strong adenine at the 3 'end of the PCR product.
Therefore, the PCR product purified by the Zymo kit was subsequently incubated for 60 min at room temperature with the tube lid open before use in the sequencing reaction and the result was significantly better, essentially perfect (see below). The problem with the first sample was probably the presence of residual ethanol from the wash buffer, which was not removed by the manufacturer's standard procedure. Additional centrifugation prior to elution (1 min), such as for the Qiagen kit, would probably resolve the situation, but is not prescribed for the Zymo kit by the manufacturer. We recommend it but it was not performed in this test.
Figure 6 - Raw data obtained by sequencing of PCR product purified by Zymo kit according to the protocol shown in Table B (upper window, 361 bp QV20 reading length) and with additional column incubation (lower window, 560 bp QV20 reading length):
Macherey-Nagel was the worst. Some analyzes did not work at all, even after the same protocol modification as in the previous case, resulting in an absolutely "dead" sequencing reaction. Slight improvement occurred after incubation of the column alone (70°C, 5 min) before loading the elution buffer, where undesirable volatile organic components could have been completely evaporated. However, the analysis was still unsatisfactory. It should also be noted that for most PCR products tested, the Macherey-Nagel kit had a lower recovery yield. In combination with a higher volume of elution buffer (compared to the NEB and Zymo kits), a poor result could also be due to the fact that although we used in the sequencing reaction quantitatively the same amount of template, its volume was higher. Thus, the Macherey-Nagel kit may not be ideal for sequencing, but may be sufficient for other applications.
Figure 7 - Raw data obtained by sequencing the PCR product purified by the Macherey-Nagel kit according to the protocol shown in Table B (upper window) and with additional column incubation (lower window):

Summary and our recommendations
Regarding the quality of the DNA template for the sequencing reaction, our experience clearly shows that the selection of the PCR purification kit is essential. If your sequencing reactions, where PCR products are used as template, do not work optimally, we can mention the following recommendations or protocol modifications in addition to changing the purification kit manufacturer:
- All ethanol from wash buffers must be completely removed from the silica membrane - by additional centrifugation before elution, by rotating the inner tube of the column during repeated centrifugation, by centrifuging the column with the lid open for better air flow through the membrane, thoroughly drying the membrane before applying the elution buffer (3-5 min at RT), by “venting” the final sample
- The ethanol in the wash buffers must be clean
- To remove other inhibitors (eg salts, detergents, agarose), multiple washes of the silica membrane with wash buffers can be performed (but this may be at the expense of the desired yield and rapid consumption of the reagents of the purification kit).
- Centrifugation with washing buffer can be performed eg after 2-5 minutes after its application on the membrane (better dissolution and removal of salts)
- Consider also the optimum volume of elution buffer - of course, the smaller the volume we choose, the higher the concentration of the final sample, but also the potential inhibitors!
Finally, we recommend our CleanSeq protocol, which includes PCR product purification, so you don't have to deal with the issues described above :-)