Better-quality sequence data at the beginning of electropherogram - Modified sequencing primers
Routinely obtained sequence electropherograms typically start with unreliable (unreadable) data just at the beginning of a sequence. Although we should theoretically read the first base after the sequencing primer (primer sequence itself cannot be detected since the synthesis and therefore labelling of a sequencing product starts right at the 3' end of the sequencing primer), there are often errors or truncations just behind the primer (usually within about 20 bases downstream of its 3' end). This causes problems if this is the region of interest or when analysing very short DNA fragments.
The cause lies in technical limitations of Sanger sequencing. First, dye terminators of the commonly used kits, “Big Dye Terminator kits”, differ in their molecular weight and charge. Therefore, these dye terminators cause significant shifts in the migration of short rather than long fragments because their contribution to the total molecular weight and charge of the sequencing product is for these short fragments higher (for longer fragment lengths these shifts are eliminated or negligible). Moreover, the capillary electrophoresis in automated genetic analysers utilizes polymers as sieving matrix and is not optimized for separation of very short and more quickly migrating DNA strands.
When there is a need to obtain high quality sequence right behind the sequencing primer, the effective way is to sequence using both forward and reverse primer or sequencing primer with additional overhang at the 5’ end (usually identical to the sequence of known universal primer). It is also possible to subclone the target DNA fragment into a vector together with using sequencing primer distant enough from the cloning site.
However, a fast, easy and reliable option we can also recommend is choosing the sequencing primer having a specific tag covalently bound to its 5’ end. This tag enables perfect reading behind any primer of your choice, only you need to make sure it is added during primer synthesis (not every primer company can do so). Nothing else is needed, you can sequence using this primer only without the need to use reverse primer or clone into plasmid as suggested above – the tag itself guarantees perfect electropherogram starting from base no. 1.
An example of this tagged-primer sequencing is shown below – there is a depiction of sequences of an identical sample when using two different primers – the standard universal primer versus the same primer but tag-modified. As you can see there is a significant improvement of read quality at the beginning of the sequence when using tag-modified primers. The pictures say it all.
Picture 1, standard primer (upper pane), tagged primer (lower pane):
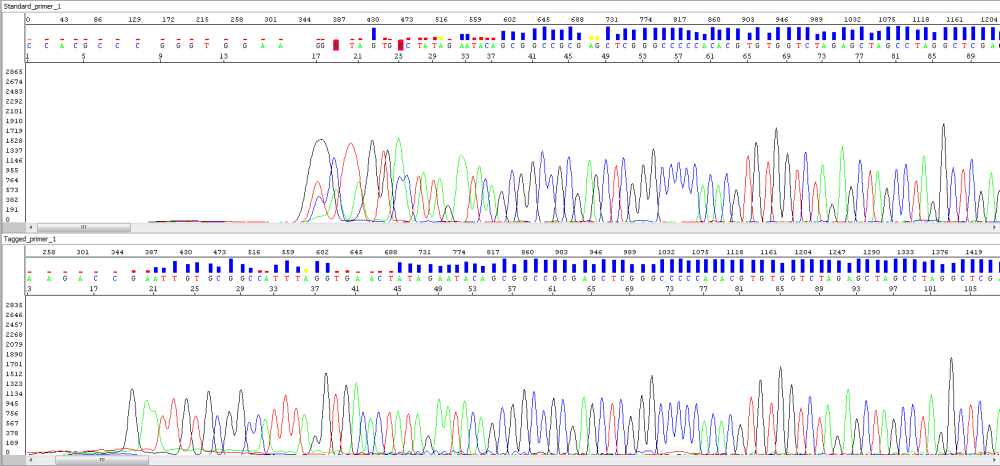
Picture 2, standard primer (upper pane), taggged primer (lower pane):
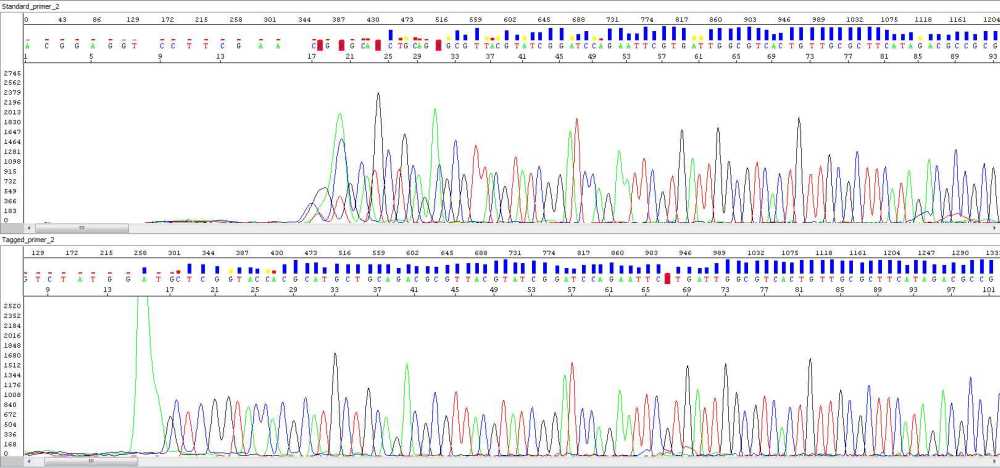
Note: Base numbers were manually shifted to visualize baseline in front of steep signal increase.
If you are interested in sequencing your samples with tag-modified primers, please contact us.
Sanger lab, info@seqme.eu