HairpinSeq – a sequencing protocol for difficult templates
Have you ever ordered sequencing analysis of a plasmid or PCR product and as a result received the DNA sequence that suddenly stops? One of the possible causes can be that in your template complex secondary structures are being formed, most often hairpins.
Some DNA fragments, especially GC-rich (usually more than 60%) or to a lesser degree AT-rich and also some vectors are prone to form intramolecular structures that are often difficult to sequence successfully. Sequencing reactions (or PCR) at standard conditions often fail due to poor denaturation - the polymerase does not pass through the region of interest or can even jump over. When looking at raw data you can see the sequence starts as expected but the signal stops abruptly (left image below) or - when the loop is slightly relaxed - the sequence starts as expected with a sudden decrease of the signal followed by little peaks of a weak intensity but still detectable (right image)
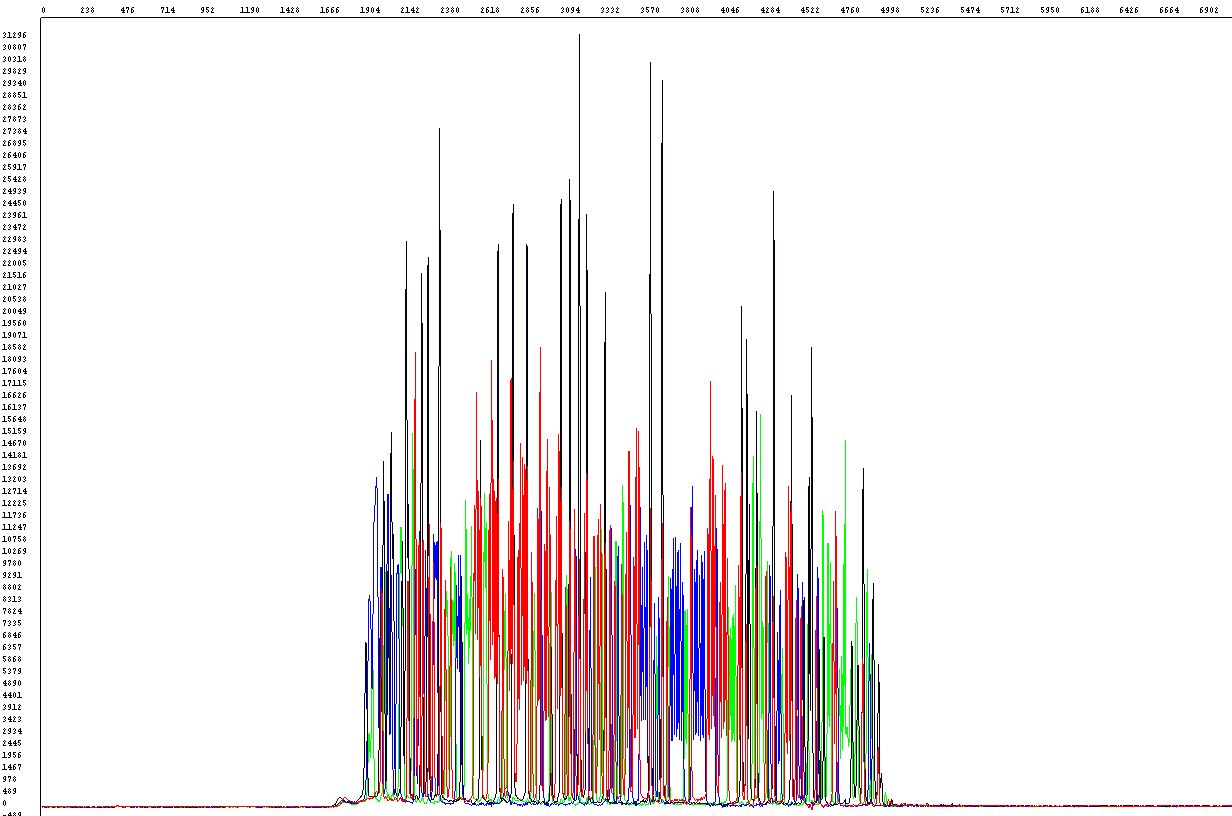
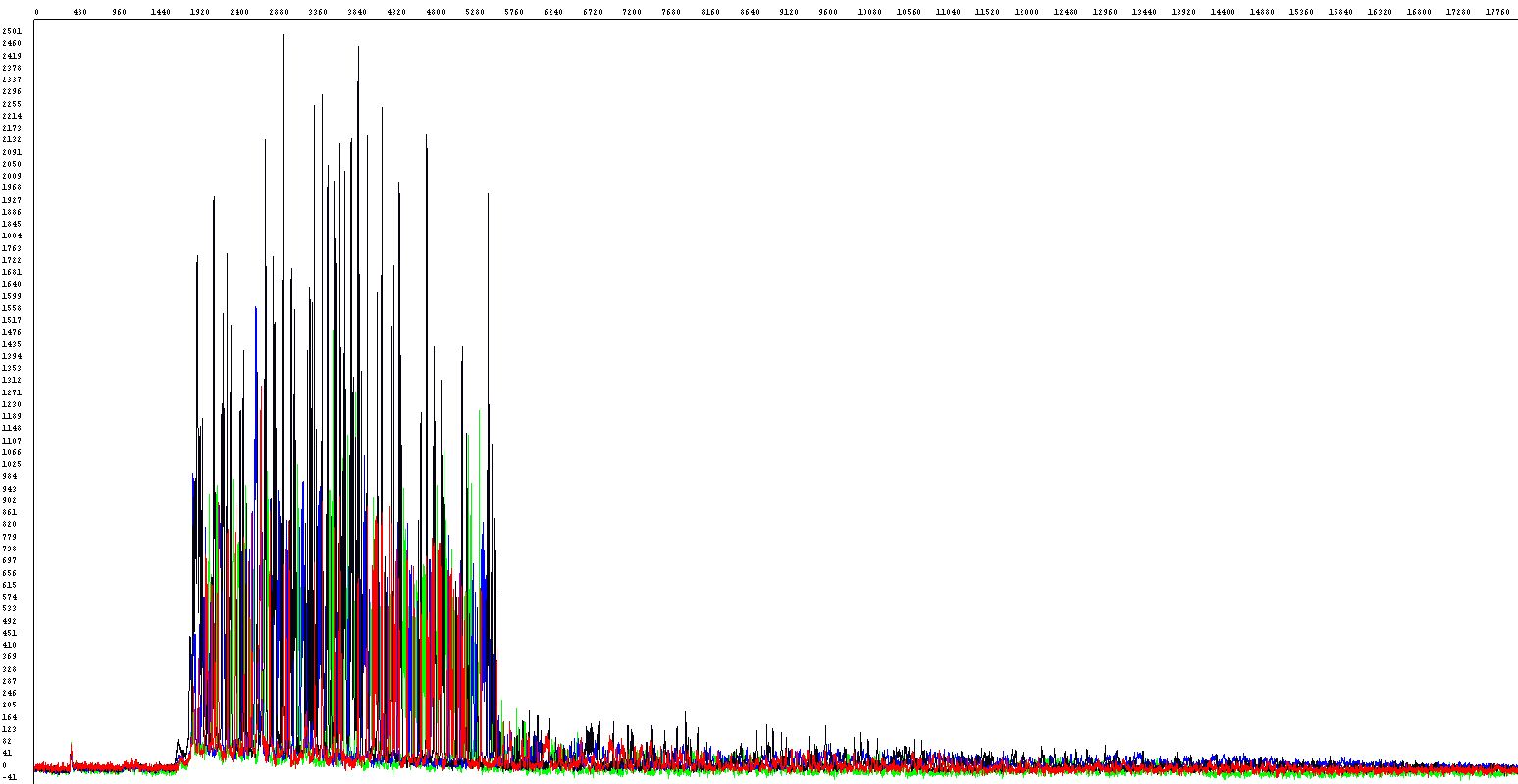
Unfortunately there is no universal and at the same time reliable method to sequence all difficult templates of this kind. There are, however, several approaches based on various additives and modified PCR procedures that can be attempted to provide satisfactory results.
First, a variety of chemical agents such as betaine, DMSO or formamide have shown the power to improve amplification and sequencing of these structures since they help to prevent the duplex formation thereby allowing the polymerase to pass through. The other possible approach is to include dGTP in the sequencing reaction. Normally its analog, dITP (forming only two hydrogen bonds with cytosine) is utilized in sequencing kits for its ability to produce high-quality data without occurrence of peak compressions when the products are separated. There are, however, some limitations when using dITP such as its inaccurate and inefficient incorporation by polymerase at higher temperatures and also in positions that are more distant from primer. Although kits including dGTP are often more capable to enable reading of difficult templates and therefore worth to try it is not recommended to use dGTP for routine applications exactly because of compressions.
It is important to mention that using additives only can be unsatisfactory without further modification of sequencing reaction conditions that help to relax the problematic structures, like higher temperature at denaturation and annealing steps or including the preincubation (heat-denaturation) step.
The sequence quality can also be greatly influenced by appropriate design of primers. Changing the distance between primer and the hairpin can sometimes lead to better results but it is definitely recommended to sequence the opposite strand too.
A considerable improvement can be reached by amplifying the difficult region and substituting 7-deaza dGTP for dGTP (usually in ratio of 1:3), ideally together with the hot-start PCR technology. 7-deaza dGTP lacks nitrogen at the 7th position of the purine ring which destabilizes formation of secondary structures by preventing alternative (Hoogsteen) base pairing without affecting the Watson-Crick base pairing.
If all options mentioned above fail it can be worth to cut the hairpin with a restriction enzyme (ideally down to fragments up to 200 bp) and subclone them – the GC content in the particular fragment is decreased which can reduce formation of the complex structures. Finally, you can try manual Maxam-Gilbert sequencing method as a last resort.
If you need to sequence difficult templates we offer the HairpinSeq protocol, our current proprietary formulation combining methods mentioned above and some others and including bidirectional sequencing using forward and reverse primer. The HairpinSeq protocol enables the readthrough of hardstops in a variety of complex difficult templates. Please note we cannot guarantee the success with every template tested since it does depend on each particular sequence.